A GENOMIC PLATFORM TO MAXIMIZE BREEDING VALUE OF GENETIC RESOURCES | 2018 Large-Scale Applied Research Project Competition (2019-2024)
Activity 3: Exploring Novel Gene Isolation and Characterization Strategies to Address Linkage Drag, Suppression and Epistasis
Activity 3.1: Suppression of Leaf Rust Resistance in Wheat
- Explore the interactions that Mediator subunits facilitate throughout the wheat genome, with a focus on Med15 homologues to identify the gene targets of SuSr-D1 and its homeologues to determine how SuSr-D1 suppresses stem rust resistance (Sr) genes.
- Identify mediator proteins involved in suppression of stem rust resistance.
- Isolate and characterize leaf and stem rust suppressor and suppressed genes.
Thatcher (ONT Assembly in place - Pozniak)
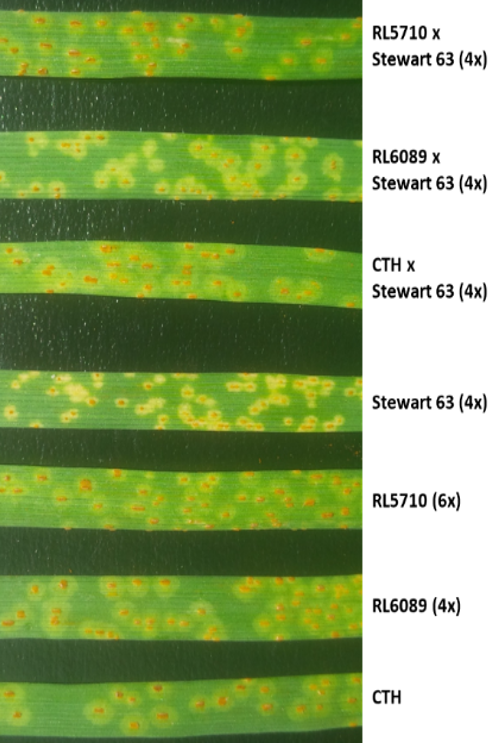
Leaf rust reactions to race BBBD. Stewart 63 and RL6089/ Stewart 63 F1 show resistant infection types
Positive Epistatic Interactions in Wheat – Leaf Rust
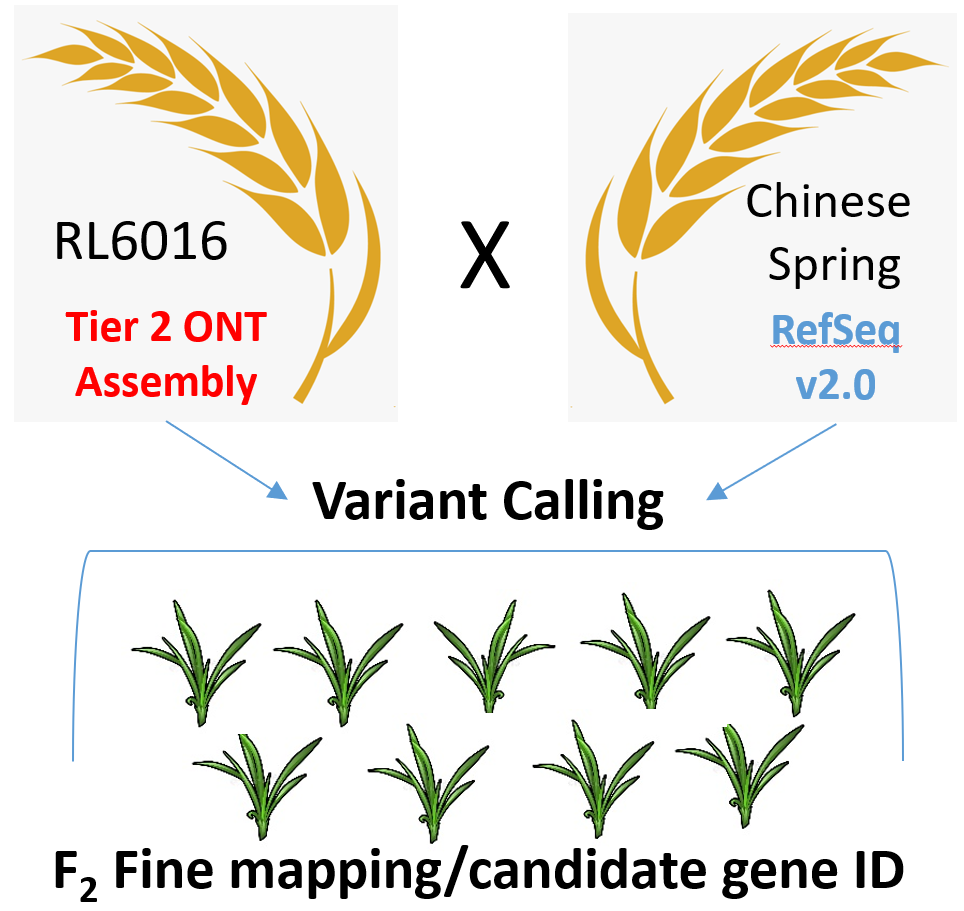
- The leaf rust resistance gene Lr2a provides excellent resistance when pyramided with Lr34, but almost no resistance in its absence. We will clone Lr2a to facilitate studies to understand the interaction with Lr34.
- RL6016 is a NIL to Thatcher (+Lr2a). An ONT assembly is available (Dr. Pozniak) from another project. A large F2 population has been generated for the cross Chinese Spring/RL6016 and is now ready for fine mapping. In addition, knock-out mutants of Lr2a are available
- The wheat stem sawfly (WSS) (Cephus cinctus Norton) is an insect pest of durum and common wheat in the northern Great Plains of North America, affecting both grain quality and yield. The most effective control of the WSS is to grow wheat cultivars with solid stems, which decreases the survival rate of insect eggs and larvae. A major gene controlling the solid-stem phenotype, SSt1, has been identified. We have cloned a candidate gene for SSt1 in durum and common wheat.
- Identification of SSt1 supressors and associated molecular markers. Once identified, the suppression factor(s) for SSt1 can be actively selected against in breeding to increase sawfly resistance in bread wheat. Unification of genomic, transcriptomic, and phenomic data into a systems biology model describing the mechanism of stem solidness and its suppression.
- The relatively narrow genetic variation for FHB resistance in elite durum wheat germplasm is as a major stumbling block for FHB resistance breeding in durum wheat. We have identified heritable FHB resistance from T. diccoccum accession TG3475151 and have localized a major QTL (QFHB-cdc.5A) on chromosome 5A that is highly stable across environments. This QTL appears to be syntenic to a stable QTL identified in cultivated emmer wheat, and is present in a few historical Canadian breeding lines.
- Improve resolution of QFHB-cdc.5A, identify QFHB-cdc.5A carriers in different genetic backgrounds and breeding materials, and develop robust DNA markers to support marker-assisted breeding.
FHB reaction of AC Morse durum wheat (susceptible) and TG3487 (resistant) 20 days after inoculation with 6 isolates of F. graminearum. Control spikes were inoculated with water

Strategy for gene mapping using a bi-parental and radiation hybrid (RH) mapping populations


