A GENOMIC PLATFORM TO MAXIMIZE BREEDING VALUE OF GENETIC RESOURCES | 2018 Large-Scale Applied Research Project Competition (2019-2024)
Activity 1: Harnessing the diversity of wheat
Proposed genome sequecning strategy
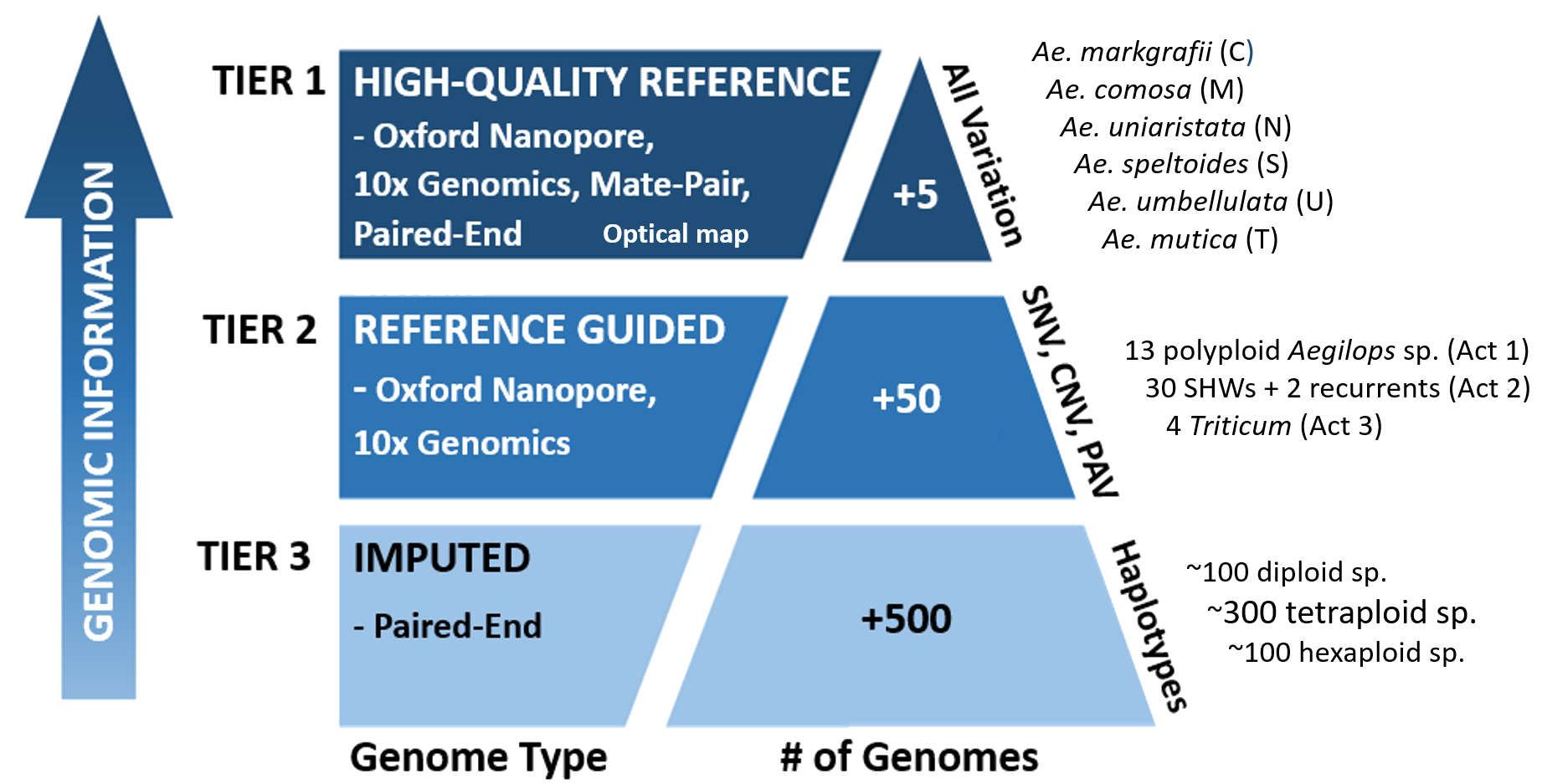 Tier 1: High-quality reference assembly
Tier 1: High-quality reference assembly
Producing high-quality de novo genome sequences for six novel genomes of Ae. markgrafii (C), Ae. comosa (M), Ae. speltoides (S), Ae. umbellulata (U), Ae. Uniaristata (N), and Ae. Mutica (T).
Genome size estimates by flow cytometry
| Species | Genome | 1C1 (Gbp) | 1C2 (Gbp) |
|---|---|---|---|
| Ae. markgrafii | C | 4.84 | 4.3±0.013 |
| Ae. comosa | M | 5.53 | 5.3±0.014 |
| Ae. speltoides | S | 5.81 | 5.5±0.005 |
| Ae. umbellulata | U | 5.38 | 5.1±0.014 |
| Ae. uniaristata | N | ||
| Ae. mutica | T |
1 Eilam, T., et al., Genome size and genome evolution in diploid Triticeae species. Genome, 2007. 50: 1029-37.
Eilam, T., et al., Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome, 2008. 51: 616-27.
2 Our estimates
Generating mid-coverage assemblies of at least 50 accessions using 30× read-depth of Oxford Nanopore, 10× read-depth of 10X Genomics and 10× read-depth of Illumina short reads technologies (Table 1.2) to enable the identification of single nucleotide variation (SNV), PAV and CNV, and a large proportion of structural variants
Mid-coverage assemblies of at least 50 accessions
| Accesion | Species | Genome | Activity | Tissue | Seed |
|---|---|---|---|---|---|
| Carberry | T. aestivum | ABD | 2.2 | √ | √ |
| Stettler | T. aestivum | ABD | 2.2 | √ | √ |
| 27 SHWs1 | T. aestivum | ABD | 1.1/1.2/2.2 | √ | √ |
| Canthatch | T. aestivum | ABD | 3.1 | √ | √ |
| RL5710 | T. aestivum | AB | 3.1 | √ | √ |
| Stewart 63 | T. durum | AB | 3.1 | √ | √ |
| DT6962 | T. durum | AB | 3.2 | √ | √ |
| D04X.00.003 | T. durum | AB | 3.2 | √ | √ |
| 13 accessions | T. turgidum ssp | AB | 1.1 | √ | √ |
| 13 accessions | Aegilops sp | Tetra/Hexa | 1.1 | √(9/13) | √(9/13) |
| 22 NAM parents | T. aestivum | ABD | 2.2 | √ | √ |
1 Includes NAM SHW parents
2 Replacement for Transcend: Assembly available based on ONT trial
- perform low-coverage sequencing (~1×) of more than 350 accessions using standard Illumina short read sequencing with the aim to define haplotypes and increase the potential to identify rare alleles.
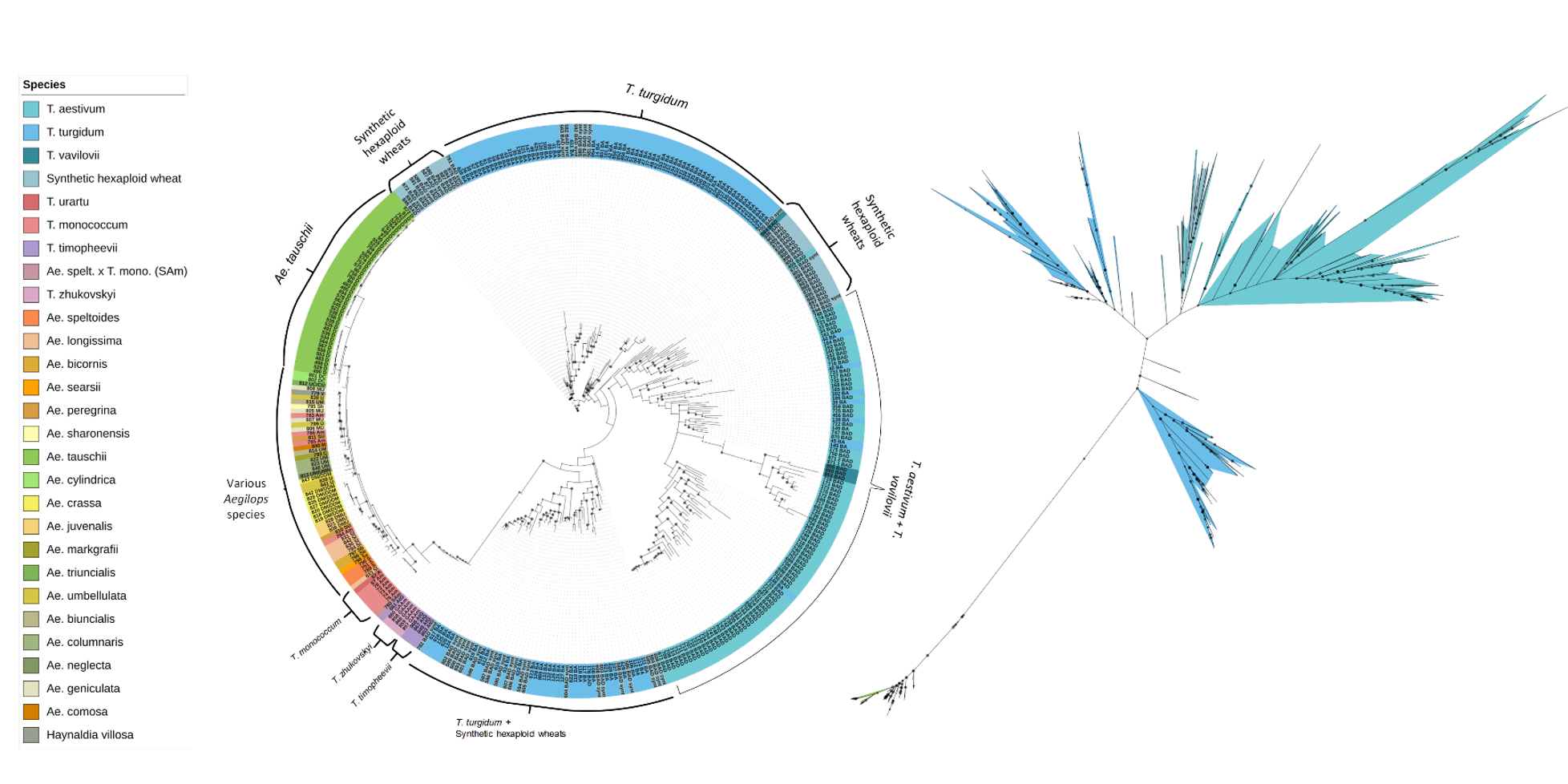
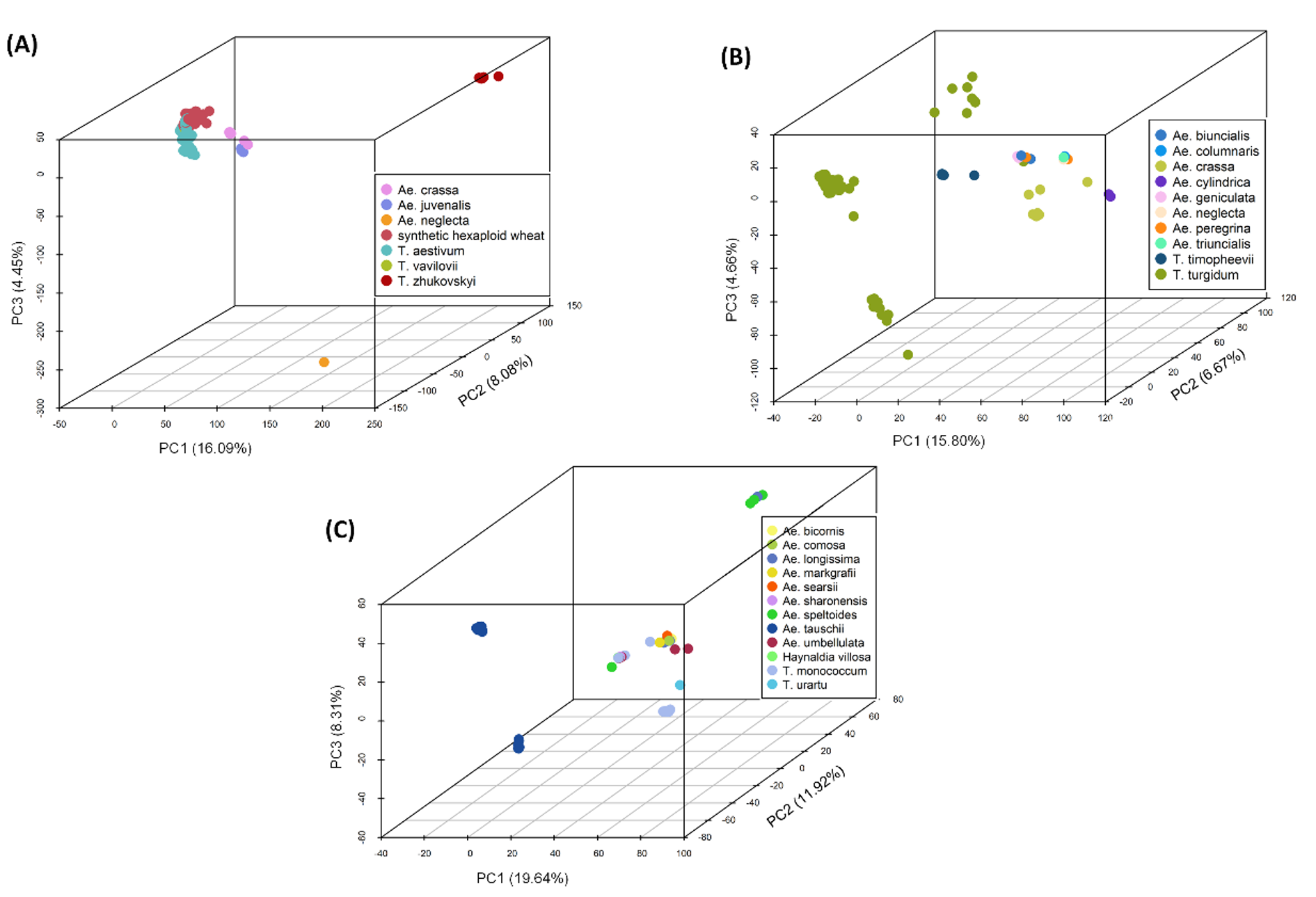
- 350 accessions of diploid, tetraploid and hexaploid from 23 Aegilops species and 4 Triticum species including many subspecies
- Selection will be based on genetic diversity, deepening the Aegilops representation and covering the subspecies of T. aestivum and T. turgidum
- Selection of tetraploid species
Genetic diversity of 350 accessions
Wild emmer wheat (T. dicoccoides)
Domesticated emmer wheat (T. dicoccum)
Durum wheat landraces (T. durum)
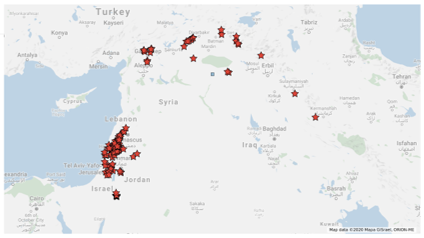
186 accessions representing Fertile Crescent region where grow WEW natural populations
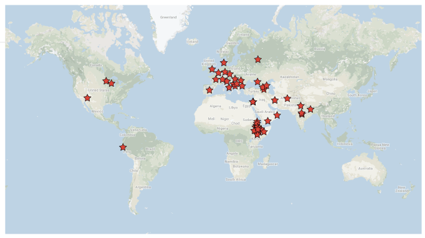
139 accessions from 29 countries
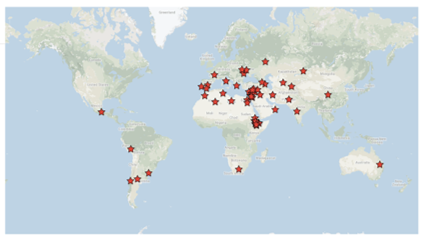
140 accessions from 39 countries